Data import formats
GelQuest imports trace data curves from the following formats:
- Trace file formats: ABI FSA format, Beckman SCF files, MegaBACE RSD files
- Gel images: BMP, TIFF, JPG, GIF, SGI, Autodesk, Targa, PCX, PCD, Portable Map, CUT, RLA, Photoshop, Paintshop, PortableNetwork, EPS
Data management and processing
- Project files are saved in XML format and contain trace data, peak data,
size standard data, etc. They are small because trace data are included
as zip-compressed CDATA sections. Easy incorporation of GelQuest samples
and projects into databases and database applications.
- Semi-automatic lane extraction from gel images:
Easy extraction from a series of image formats.
Full UNDO functionality to the image import steps.
Smile correction. Correction of distorted gel runs.
- Supports up to 5-color channel fingerprinting systems
- Trace signal processing: smoothing, baselining
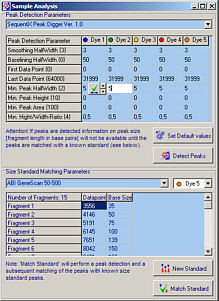
- Peak detection with realtime preview:
Advanced SequentiX peak digger algorithm.
Many peak detection and filtering parameters.
- Fragment sizing: see below
- Allows for the comparison (binning) of fragments even if no size standard lane is available
- Create/Edit user-defined size standard templates
- Comes with predefined standard templates
Click image to enlarge
Sample management
- Detailed information on each sample
- Renaming (edit sample names)
- Rearrange samples within a project by drag and drop
- Remove/Add samples from the project
- Analyse individual samples
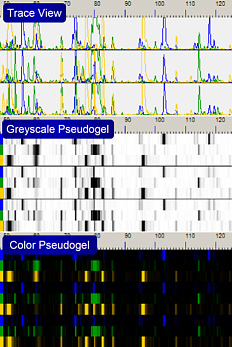
Data Visualisation (graphics)
- Select the color channels to display
- Display as curves, color or greyscale pseudogel images
- Show/Hide peak locations
- Toggle between raw and processed data
- Scale by base size or datapoints
- Adjust all scaling parameters
- Various zoom functions (zoom in, zoom out, zoom in range)
- For each peak detailed information is available on the fly
- Adjust the scaling parameters, e.g. normalization of RFU values to whole run or to the visible region.
- Peak detail window with peak height, width, area, height-to-width-ratio, datapoint, size
Click image to enlarge
|
|
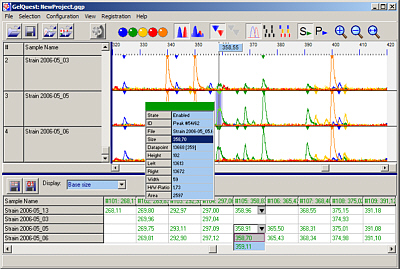
Easy handling
This is a screenshot of GelQuest's main window showing the trace
panel with peak details in the upper region and the hyperbin table
in the lower part of the window:
Click image to enlarge
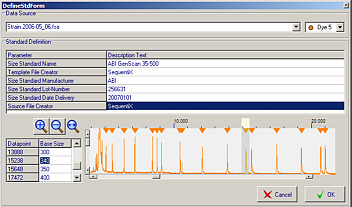
Size standard definition
Use your own size standards and generate size standard template files that can be used to size any of your samples.
Automatic size standard matching for fragments with size standard templates.
Local Southern algorithm for exact peak sizing.
Peaks occuring above the highest and below the smallest size standard peak are sized by linear regression.
Allows sizing and hyperbinning of fragments with any length (also suitable for pulse field gel electrophoresis data).
Allows for the comparison/binning of fragments even if no size standard lane is available.
Definition of user-defined size standard templates from samples.
Comes with several predefined standard templates.
Click image to enlarge
Hyperbins (Superbins)
- Automatic hyperbin detection and on-the-fly visualisation of corresponding bins in a hyperbin-table
- Lucid presentation of hyperbins by base sizes, heights, areas, area-to-height-ratios or as a 0/1 matrix
- Include or exclude sample peaks by clicking with the right mouse button on a trace peak or the hyperbin table
- Adjust hyperbin detection parameters and filter peaks by height, area, height to width ratio or other parameters
- The maximum hyperbin width is automatically adjusted to the maximum base size of peaks divided by 2.
- Filter by channels
- Advanced local maximum detection algorithm
- Bin = a single peak of a single sample
- Superbin = a container on sample level with a defined width (e.g. 1 base) containing bins (peaks) of a single sample
- Hyperbin = a container on project level containing bins and superbins of serveral samples
- Export of hyperbins for spread sheet applications in tab-delimited tables
- Export hypberbins as 01 matrix files for cluster analyses (supported formats: Paup NEXUS, Phylip NEWICK and FASTA)
- Start add-on analysis tool ClusterVis to perform cluster analysis or principal coordinate analysis (PCoA).
Export Formats
- Trace files (FSA format)
- Trace data (raw or processed) scaled by base sizes or datapoints into tab-delimited tables for spead sheeth applications
- Peak data in tab-delimited files
- Hyperbins as a table
- 01-matrix files for cluster analyses (supported formats: Paup NEXUS, Phylip NEWICK and FASTA)
ClusterVis (Add-On Software)
- Cluster-Analysis of binary data
- 01 matrix import, export, editing
- 01 matrix import, export, editing
- Takes over 01-matrices directly from GelQuest
- Distance matrix import, export, copy
- Calculation of distance matrices. Over 50 distance measures are
already implemented. 26 measures are evaluated and available in
the current release.
- Cluster analysis using UPGMA and Neighbor-Joining algorithms
- Visualization, saving and copying of cluster trees: radial tree, cladogram, elliptogram, phylogram
Available 12/2007:
- Testing of robustness of trees: Bootstrapping, Jack-knifing
- Principal coordinates analysis (PCoA)
|
|




